Genetic Variation of PD-1 is Associated with The Development of Hepatocellular Carcinoma in Patients with Chronic Hepatitis C Infection
Fatima Zahra Jadid 1,2*, Hajar Chihab 1, Hanane Salih Alj2, Raouia Elfihry1, Imane Zaidane1, Wafaa Badre3, Mohammed Tahiri3, Isabel Chemin4, Pascal Pineau5, Sayeh Ezzikouri1, Soumaya Benjelloun1
1 Virology Unit, Viral Hepatitis Laboratory, Institut Pasteur du Maroc, Casablanca, Morocco.
2 Laboratoire de Biologie et Santé, URAC 34, Faculté des Sciences Ben M’Sick, Université Hassan II de Casablanca, Morocco.
3 Service d’Hépato-Gastro-Entérologie, CHU Ibn Rochd, Casablanca, Morocco.
4 INSERM U1052, CNRS UMR5286, Centre de Recherche en Cancérologie de Lyon Université Claude Bernard, Lyon, France.
5 Unité Organisation Nucléaire et Oncogenèse, INSERM U993, Institut Pasteur, Paris, France.
*Corresponding Author:Jadid Fatima Zahra, Virology Unit, Institut Pasteur du Maroc, 1 Place Louis Pasteur, 20360 Casablanca, Morocco, Tel: +212 5 27016076/ 5 22434450; Fax: +212 5 22260957; E-mail:jadid.fz@gmail.com
Citation: Fatima Zahra Jadid, Hajar Chihab, Hanane Salih Alj, Raouia Elfihry, Imane Zaidane, et al. (2022) Genetic variation of PD-1 is associated with the development of hepatocellular carcinoma in patients with chronic hepatitis C infection. Gastroenterol Hepatol J 5: 127.
Received: October 26, 2022; Accepted: November 02, 2022; Published: November 05, 2022.
Copyright: © 2022 Fatima Zahra Jadid, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Introduction & Objectives: Hepatitis C virus persistence and pathobiology result from the interplay between viral replication and host immune responses. Programmed cell death-1 (PD-1) is an important immune effector with co-inhibitory activity involved in the progression of chronic viral infections. Our aim was to investigate the influence of the functional single nucleotide polymorphism (rs10204525) in the 3’-UTR of PD-1 and PD-1 mRNA expression on the outcomes of hepatitis C virus infection including disease progression in Moroccan patients.
Patients & Methods: A total of 200 healthy controls, 101 spontaneous resolved HCV patients and 300 chronic HCV subjects (95 patients with mild liver disease, 131 individuals with advanced liver disease and 74 patients with hepatocellular carcinoma, HCC) were enrolled in this study and genotyped for rs10204525 using TaqMan allelic discrimination. PD-1 mRNA expression in peripheral blood nuclear cells was determined by qRT-PCR. PD-1 mRNA expression in peripheral blood nuclear cells was determined by qRT-PCR.
Results: Multivariate logistic regression analysis showed the significant association of rs10204525 with progression liver disease (OR= 1.748, 95% CI = 1.034–2.955, P = 0.036). In addition, the T allele was related to an increased risk of HCC among patients with chronic HCV infection (OR= 1.528, 95% CI = 1.022–3.284, P = 0.038). In addition, PD-1 mRNA was overexpressed in chronic HCV infected patients with cirrhosis and hepatocellular carcinoma when compared to mild fibrosis group (P=0.0002) and the expression was even more pronounced when compared to HCV resolved group (P<0.0001).
Conclusion: These findings underline the importance of the functional polymorphism in PD-1 in the installation of HCV infection and its subsequent contribution to disease progression including the development of HCC.
Keywords: Chronic HCV Infection, Programmed Cell Death-1, HCC, PD-1 Polymorphism
Abbreviations
PD-1: Programmed Cell Death-1, IgSF: Immunoglobulin Super Family, HCV: Hepatitis C Virus, HCC: Hepatocellular Carcinoma, CTL: Cytotoxic T Lymphocytes, AdLD: Advanced Liver Diseases, mLD: Mild Liver Diseases, PBMCs: Peripheral Blood Mononuclear Cells, GAPDH: Human Glyceraldehyde-3-Phosphate Dehydrogenase, MAF: Minor Allele Frequency
Introduction
Chronic hepatitis C (CHC) represents a serious global health burden with 170 million people chronically infected worldwide [1] and leads to over 350,000 deaths each year [2]. Hepatitis C virus (HCV) exhibits a remarkable propensity to cause hepatic fibrosis, cirrhosis, hepatocellular carcinoma (HCC) and liver-related mortality [3]. The nucleocapsid core protein is the first protein synthesized following HCV viral infection, and is well conserved in different HCV genotypes. It has been demonstrated that HCV core protein inhibits T-cell proliferation and promotes B-cell activation through differential regulation of PD-1 signaling in vitro [4]. Additionally, HCV infection is associated with T-cell dysfunction and increased expression of PD-1 in T cell dysfunction [5].
Programmed cell death 1 (PD-1) is one of the latest immune regulatory genes that belongs to the immunoglobulin superfamily (IgSF). PD-1 gene is located on 2q37.3 and expressed on activated T and B lymphocytes, natural killer cells and monocytes. PD-1 protein is a transmembrane glycoprotein of the CD28 immunoglobulin superfamily and contains an immunologic receptor tyrosine-based inhibitory motif [6, 7]. With engagement to its ligands, PD-L1 (B7-H1) and PD-L2 (B7-DC), PD-1 can inhibit cytokine secretion by downregulate T cells proliferation activity, enhance peripheral tolerance and induce T cell apoptosis [8, 9]. PD-1 can be induced by viral infection and its expression decreased after efficient antiviral therapies [10]. PD-1 is involved in immune disruption and viral persistence during chronic HCV infection [11] and is an important inhibitory receptor that downregulates T-cell function [12]. Studies focusing on CHC have shown that upregulation of PD-1 affects HCV specific CD8+ T cells in the liver and peripheral blood during chronic HCV infection [11, 13]. Moreover, several lines of evidence have been reported on the importance of CD8+ T cells during HCC [14, 15]. Furthermore, Shi et al demonstrated that PD-1 and PD-L1 upregulation promotes CD8+ T cell apoptosis recurrence in patients with HCC [16].
In addition to the viral and immune factors, individual susceptibility factor has been identified to be one of the most important factors affecting the prognosis of the disease related to the HCV chronic infection. Recently, PD-1 polymorphisms were shown to be associated with the susceptibility and outcomes of HCV infection [17] and also with the susceptibility and disease progression in chronic hepatitis B virus (HBV) infection [18].
Likewise, recent reports have highlighted that PD-1 variants are associated with susceptibility to several types of cancer, such as breast cancer [19, 20], colon cancer [21], gastric cancer [22, 23], esophageal cancer [24] and hepatocellular carcinoma [18]. However, the association between PD-1 polymorphisms and progression of chronic hepatitis C and HCC development was unclear. So far, the role played by rs10204525 in chronic infection with HCV has been studied only in patients of Han ethnicity. To extend our knowledge about this potentially crucial polymorphism in North Africa, we investigated the influence of rs10204525 and mRNA expression on the outcome of chronic HCV infection in a Moroccan population.
Patients and Methods
Study Subjects
A cohort of 601 Moroccan subjects including 226 patients with chronic HCV infection, 101 patients spontaneously resolved from HCV infection and 200 healthy controls were enrolled in this study at the Medical Center of Biology at Pasteur Institute of Morocco and Service of Medicine B CHU Ibn Rochd Hospital, Casablanca, from September 2013 to September 2016. All individuals have given informed consent and the study was approved by the Ethics Committee of the Faculty of Medicine of Casablanca.
The persistent infection group was positive for anti-hepatitis C virus (anti-HCV) antibodies and HCV RNA determined by a quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) for at least six months. Among them, 95 had mild chronic hepatitis C (mCHC, patients with minimal fibrosis score F0 and F1-F2), 131 with HCV-related-AdLD (patients with advanced fibrosis F3-F4) and 74 with HCC. Spontaneous viral clearance was defined as anti-HCV antibodies positive and undetectable HCV RNA. The healthy controls had no medical history of any liver disease and they were negative for viral hepatitis markers and had normal serum levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST). Subjects co-infected with any other virus were excluded.
Serological markers for HBsAg, anti-HCV and anti-HIV were tested with commercially available kits (Abbott Architect i2000SR Analyzer (Abbott Laboratories, Abbott Park, IL, USA). All participants of this study gave their informed consent and in person interview was conducted using a structured questionnaire to get information about demographic data, medical history, lifestyle and other characteristics. The peripheral blood from the study subjects was collected into EDTA-containing tubes.
DNA extraction and PD-1 polymorphism genotyping
Peripheral blood samples were collected and stored at -20°C. The genomic DNA was carefully extracted from Peripheral Blood Mononuclear Cells (PBMC) of all subjects. The PD-1 rs10204525 A>G polymorphism was genotyped with primers and fluorescence dual color hybridization probes specific for PD-1 polymorphism (Applied Biosystems; assay ID C_172862_10) using the LightCycler® 480 Real-Time PCR System (Roche diagnostics, Manheim, Germany).
RNA extraction and PD-1 mRNA expression
Total RNA was isolated from PBMCs using the TRIzol reagent (Invitrogen; USA). Total RNA was stored at −80°C until use. cDNA was synthesized from 10µl of extracted total genomic RNA by using MMLV Reverse Transcriptase (Bioline, London, UK) and Random Hexamer primer Mix, according to manufacturer’s instructions. Quantitative real-time PCR on cDNA were performed in triplicates using SYBR Green PCR Master Mix (Applied Biosystems) and PD-1 primers sequences described previously [25]. Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control and Ct was calculated from the differences in the mean Ct between the PD-1 gene and the internal controls. Primers used for GAPDH have been published previously [26].
Statistical Analysis
Statistical analysis was carried out using SPSS 21.0 software (SPSS, Chicago, Illinois, USA). Age was presented as the mean ± standard deviation (SD) and we used Student’s t-test to determine the differences for continuous variables between groups. Chi-square test (X2) was used to compare the categorical variables and the genotypes distributions between groups. Hardy-Weinberg equilibrium was assessed by a goodness-of-fit χ2 test with 1 degree of freedom to compare observed and expected genotype frequencies. Multivariate logistic was conducted to obtain the odds ratios (OR) and their 95% confidence intervals (CI) for the relationship of PD-1 rs10204525 T>C polymorphism with hepatic fibrosis progression and hepatocellular carcinoma risk. The statistical significance was defined as (P<0.05).
Results
Demographic and clinical characteristics of the study subjects
The baseline of the general characteristics of the 300 chronic HCV patients, 101 spontaneous resolved patients and 200 healthy controls enrolled in this study are summarized in (Table 1). The age and gender between groups were not statistically different (P>0.05). Although, significant differences were observed in the distribution of ALT and AST between CHC group and healthy controls (P<0.0001). Likewise, total cholesterol, LDL-cholesterol and triglyceride levels were very significantly decreased in the CHC group as compared to the healthy subjects (P<0.0001). There was no difference in the distribution of parameters between CHC group and spontaneous resolved patients.
Effect of PD-1 polymorphism on the HCV infection and resolution
The genotype distributions of rs10204525 among chronic HCV patients, spontaneous resolved patients and healthy controls are shown in Table 2. Multivariate logistic regression analysis showed no significant associations of PD-1 polymorphism with HCV spontaneous clearance were observed between HCV chronically infected patients and subjects who spontaneously resolved the infection.
The impact of PD-1 polymorphism on progression liver disease and hepatocellular carcinoma
In order to analyze the effect of PD-1 polymorphism on liver disease progression and hepatocellular carcinoma in the Moroccan population, we compared genotypes distributions for rs10204525 in 95 patients with mCHC, 131 individuals with AdLD and 74 subjects with HCC. Among 95 of individuals with mild fibrosis, 59 (62.29%) were homozygous for the wild type CC, 30 (31.58%) were heterozygous and 6 (6.31%) were homozygous for TT. In Advanced fibrosis and HCC group, genotype distribution was identified as, CC homozygous in 99 (48.29%) subjects, heterozygous in 88 (42.93%) and homozygous TT in18 (8.78%). The genotype distribution of this SNP was in accordance with the Hardy-Weinberg equilibrium (P>0.05). Multivariate logistic regression analysis showed the significant association of rs10204525 with progression liver disease (OR= 1.748, 95% CI = 1.034–2.955, P = 0.036). In addition, the T allele was related to an increased risk of HCC among patients with chronic HCV infection (OR= 1.528, 95% CI = 1.022–3.284, P = 0.038) (Table 3). These findings underline the importance of the functional polymorphism in PD-1 in the installation of HCV infection and its subsequent contribution to disease progression including the development of HCC.
Table 1: Demographic, clinical and biochemical characteristics of the subjects included.
|
Characteristics |
Persistently infected |
Spontaneous clearance |
Healthy controls |
|
|
patients (n=300) |
subjects (n=101) |
(n=200) |
|
Mean age ± SD, y |
63.12 ± 12.11 a* |
59.14 ± 13.75 |
56.11 ± 13.77 |
|
Gender N (%) Male |
109 (36.33) ns |
42 (41.58) |
67 (33.5) |
|
Female |
191 (63.66) |
59 (58.42) |
133 (66.5) |
|
Mean ± SD ALT (IU/L) |
80.01 ± 55.87 a* |
46.44 ± 56.24 |
35.33 ± 21.52 |
|
AST (IU/L) |
68.28 ± 49.19 a* | 34.98 ± 21.58 | 29.35 ± 16.39 |
|
Mean bilirubin (µmol/L) |
15.19 ± 6.08 | 15.16 ± 11.17 |
Na |
|
Mean creatinine (mmol/L) |
108.54 ± 199.72 | 82.34 ± 34.55 |
Na |
|
Fasting serum glucose (g/L) |
1.06 ± 0.41 a | 1.23 ± 0.54 | 0.95 ± 0.19 |
|
Total cholesterol (g/L) |
1.53 ± 0.34 a* | 1.72 ± 0.37 | 1.91 ± 0.38 |
|
Triglycerides (g/L) |
1.05 ± 0.38 a* | 1.37 ± 1.33 | 1.31 ± 0.68 |
|
HDL-cholesterol (g/L) |
0.52 ± 0.17 | 0.48 ± 0.10 | 0.54 ± 0.37 |
|
LDL-cholesterol (g/L) |
0.82 ± 0.53 a* | 0.94 ± 0.35 | 1.12 ± 0.36 |
|
Median viral load (IU/ml) |
2.8 E+06 |
|
- |
|
|
[0.9 E+03-64.5 E+06] |
|
|
|
Viral genotypes (%) |
|
|
|
|
Genotype 1 |
59.90 |
- |
- |
|
Genotype 2 |
39.06 |
- |
- |
|
Genotype 3 |
39.06 |
- |
- |
|
Genotype 4 |
0.52 |
- |
- |
|
mCHC |
95 |
|
|
|
AdLD |
131 |
|
|
|
HCC |
74 |
|
|
ALT: Alanine aminotransferase
AST: Aspartate aminotransferase
SD: Standard deviation
aSignificant values (P < 0.05) when compared to the healthy controls
a*Significant values (P < 0.0001) when compared to the healthy controls
Na: Non applicable
Table 2: Effect of PD-1 polymorphism on the HCV infection susceptibility and resolution.
|
|
Healthy controls (n=200) |
Persistent infection (n=300) |
Subjects with spontaneous clearance |
Healthy controls vs. Subjects Persistent infection OR (95% |
P-value |
Subjects with SpC vs. Subjects with persistent infection |
P-value |
||
|
|
|
(SpC) (n = 101) |
CI) |
|
OR (95% CI) |
|
|||
|
PD (rs10204525) CC |
88 (44%) |
158 (52.67%) |
56 (55.45%) |
1 (Reference) |
- |
1 (Reference) |
- |
||
|
CT |
89 (44.5%) |
118 (39.33%) |
32 (31.68%) |
0.11653 | 0.11653 | 1.307 (0.796-2.145) |
|
||
|
TT |
23 (11.5%) |
24 (8%) |
13 (12.87%) |
0.581 (0.310-1.090) | 0.08826 | 0.654 (0.312-1.372) | 0.25930 | ||
|
C allele |
0.662 ± 0.023 | 0.723 ± 0.018 | 0.713 ± 0.035 |
1 (Reference) |
- |
1 (Reference) |
- |
||
|
T allele |
|||||||||
|
Dominant |
|
158/142 |
56/45 |
0.706 (0.493-1.012) | 0.05756 | 1.118 (0.711-1.760) | |||
|
model Recessive model |
|
276/24 |
88/13 |
|
|
|
Table 3: The impact of PD-1 polymorphism on progression liver disease and hepatocellular carcinoma.
|
Healthy controls (n = 200) |
Mild fibrosis group (n = 95) |
Advanced fibrosis and HCC group (n = 205) |
Healthy controls vs. Advanced fibrosis and HCC group OR (95% CI) |
P-value |
Mild fibrosis group vs. Advanced HCC Patients OR (95% CI) |
P-value |
|
PD-1 (rs1020452) CC 88(44%) |
59 (62.11%) |
99 (48.29%) |
1 (Reference) |
- |
1 (Reference) |
- |
|
CT 89(44.5%) |
30 (31.58%) |
88 (42.93%) |
0.879 (0.582-1.326) | 0.538530 | 1.748 (1.034-2.955) | 0.03612 |
|
TT 23(11.5%) |
6 (6.31%) |
18 (8.78%) |
0.696 (0.352-1.374) | 0.29434 | 1.788 (0.672-4.757) | 0.23971 |
|
Callele 0.662 ±0.023 |
0.779 ± 0.031 | 0.698 ± 0.022 |
1 (Reference) |
- |
1 (Reference) |
- |
|
Tallele 0.338 ±0.023 |
0.221 ± 0.031 | 0.302 ± 0.022 | 0.851 (0.633-1.144 | 0.28475 | 1.528 (1.022-2.284) | 0.03817 |
|
Dominant |
59/36 |
99/106 |
0.841 (0.569-1.244) | 0.38629 | 1.755 (1.068-2.884) | 0.02582 |
|
model Recessive |
89/6 |
187/18 |
1.350 (0.705-2.586) |
0.36434 |
0.700 (0.269-1.825) |
0.46418 |
PD-1 rs10204525 polymorphism and clinical markers in patients with chronic HCV infection Demographic, biochemical and viral data were analyzed according to PD-1 rs10204525 polymorphism (Table 4). Individuals with CT genotype have higher AST levels compared to subjects with CC and TT genotype (P<0.005). Though, other clinic-pathological markers did not reveal any significant difference (P>0.05, Table 4).
Table 4: PD-1 rs10204525 polymorphism and clinical markers in patients with chronic HCV infection.
|
|
CC genotype (n=125) |
CT genotype (n=82) |
TT genotype (n=19) |
|
Mean age ± SD, y |
60.94 ± 12.26 |
61.44 ± 11.38 ns |
61.89 ± 14.65 ns |
|
Gender (%) Male |
39 (31.2) |
21 (25.61) ns |
7 (36.84) ns |
|
Female |
86 (68.8) |
61 (74.39) |
12 (63.16) |
|
Mean ± SD |
|
||
|
ALT (IU/L) |
74.01 ± 51.21 | 83.50 ± 62.51 ns | 87.00 ± 55.82 ns |
|
AST (IU/L) |
57.40 ± 33.98 | 78.54 ± 62.62 ** | 59.11 ± 23.05 ns |
|
Mean bilirubin (µmol/L) |
15.53 ± 6.28 | 15.00 ± 6.17 ns | 14.27 ± 4.25 ns |
|
Mean creatinine (mmol/L) |
105 ± 192.71 |
105 ± 192.71 | 100.50 ± 221.52 ns |
|
HDL-cholesterol (g/L) |
0.54 ± 0.18 | 0.50 ± 0.16 ns | 0.54 ± 0.13 ns |
|
LDL-cholesterol (g/L) |
0.82 ± 0.65 | 0.85 ± 0.31 ns | 0.73 ± 0.19 ns |
|
Median viral load (IU/ml) |
2.4 E+063. |
3.2 E+06 ns | 2.8 E+06 ns |
|
|
[0.9 E+03-31.8 E+06] |
[0.9 E+03-64.5 E+06] |
[9.6 E+03-9.4 E+06] |
|
Viral genotypes n (%) Genotype 1 |
85 (68) |
51 (62.2) ns |
14 (73.7) ns |
|
Genotype 2 |
38 (30.4) |
31 (37.8) |
5 (26.3) |
|
Genotype 3 |
1 (0.8) |
- |
- |
|
Genotype 4 |
1 (0.8) |
- |
- |
|
mCHC |
59 |
30 ns |
6 ns |
|
AdLD |
66 |
52 |
13 |
ALT: Alanine aminotransferase AST: Aspartate aminotransferase SD: Standard deviation *Significant values (P < 0.05) when compared to the CC genotype ** Significant values (P < 0.005) when compared to the CC genotype *** Significant values (P < 0.0001) when compared to the CC genotype Na: Non applicable
Analysis of PD-1 mRNA expression according to rs10204525 genotypes and clinical parameters
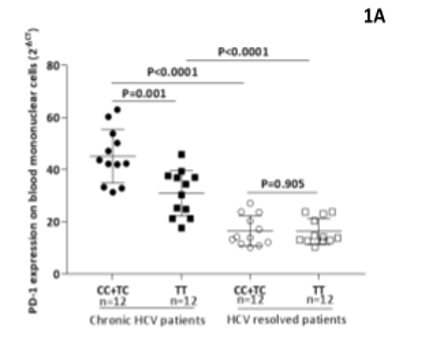
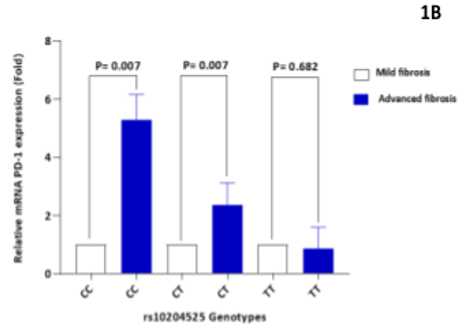
The PD-1 mRNA expression levels were detected in peripheral mononuclear cells of chronic HCV patients and spontaneous resolved individuals according to rs10204525 genotypes. As shown in Figure 1A, the expression of PD-1 mRNA in chronic HCV patients was significantly higher than that in HCV resolved patients (P<0.0001). Among chronic HCV patients, the expression of PD-1 mRNA was significantly higher in patients with PD-1 rs10204525 CC or TC genotype than those carrying the TT genotype (P=0.001). No significant difference was observed in PD-1 rs10204525 genotypes among HCV resolved patients (Figure 1A). To evaluate the effect of PD-1 gene expression in HCV-infected patients at different stages of liver disease. Taking as baseline expression levels in mCHC patients, a statistically significant 6-fold upregulation of PD-1 expression was observed in the AdLD group carrying CC genotype (P=0.007, Figure 1B).
Figure 1: PD-1 expression on blood mononuclear cells in chronic HCV patients and HCV resolved patients according to rs10204525 genotypes. Mean ± SD in different groups are shown. The horizontal bars among the symbols indicate mean and 95% CI values in each group. P-values were determined by independent t-test (1A). PD-1 relative expression levels according to rs10204525 genotypes in HCV infected patients with advanced fibrosis and mild fibrosis patients (1B).
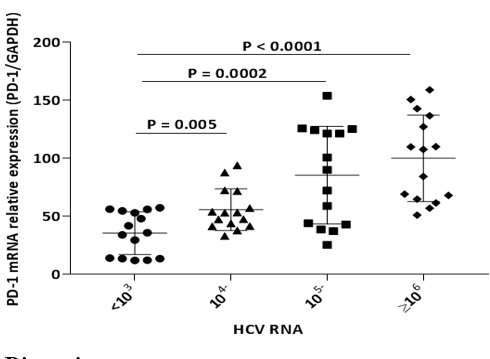
In addition, PD-1 mRNA was overexpressed in chronic HCV infected patients with cirrhosis and hepatocellular carcinoma when compared to mild fibrosis group (P=0.0002) and the expression was even more pronounced when compared to HCV resolved group (P<0.0001, Figure 2). Overall, differences in PD-1 expression were found to be associated with liver disease progression. Likewise, similar analysis according to HCV RNA was shown that PD-1 mRNA relative expression levels was significantly different among patients with HCV RNA levels (IU/mL) of <103, 104~, 105~ and ≥106 (P<0.0001), (Figure 3). The analysis of PD-1 gene expression according to age, sex and metabolic parameters of the population did not show any significant differences (data not shown).
Figure 2: PD-1 mRNA levels in patients with different clinical diseases of chronic hepatitis C infection and HCV resolved patients.
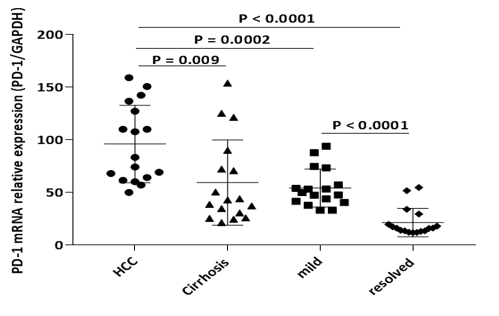
Figure 3: PD-1 mRNA expression in patients with chronic HCV infection of different HCV RNA levels.
Discussion Chronic HCV infection is a major cause of cirrhosis and HCC [27, 28]. It is known that genetic variation of the host, viral and environmental factors are modulating disease progression [29]. In this study, we analyzed the impact of rs10204525 and mRNA expression on and outcome of chronic HCV infection. The PD-1 protein plays a role in the progression to chronic viral infection [30]. It has been reported in various studies that an elevated expression of PD-1 gene on peripheral lymphocytes is strongly associated with the dysfunction of immune response in chronic HCV infection and HCV-related HCC [31, 32]. Additionally, it has described that PD-1 attenuated the immunoregulation of T cells, which finally resulted in chronic viral infection [9].
In the present study, a significant relationship between PD-1 expression and HCV infection progression was found. PD-1 mRNA was highly expressed in patients with cirrhosis and HCC and this expression was significantly associated with the CC genotype of rs10204525 polymorphism. In addition, a significant association between increased PD-1 expression on PBMCs and high HCV RNA load levels of HCV viral replication was revealed in this study. This finding supports previous studies that the increased PD-1 mRNA levels were correlated with higher HCV RNA levels, which represents an indicator of active HCV viral replication [31, 33]. With respect to the association of PD-1 mRNA levels with rs10204525 variant, this study showed that PD-1 mRNA expressions were sequentially increased from PD-1 rs10204525 genotypes TT, CT to CC (p=0.001). This result seems in line with recent studies conducted in the Moroccan HBV infected subjects [34] and in treated HIV-1-infected Moroccan subjects (p< 0.005) indicating the same observations [35]. However, at odds with our findings, Xiao and colleagues found a decrease in PD-1 mRNA levels from PD1 rs10204525 TT, CT, to CC genotype in patients with CHC [17]. The same result was observed also in CHB [25]. Interestingly, the significant increase of PD-1 mRNA levels in patients carrying TT genotype was the hallmark of chronic hepatitis in Eastern Asian populations [17, 25]. In the current study, we found that T allele was associated with progression of liver fibrosis and related to HCC among patients with chronic HCV infection. This result was in opposite with earlier studies showing that T allele was associated with spontaneous clearance [17]. This discrepancy between results found in studies conducted on subjects with different ethnicity may be related to the genetic background or to allele frequency differences between populations.
In conclusion, this study showed that patients with chronic infection had significantly elevated levels of PD-1 mRNA expression that were correlated with clinical diseases and HCV viral replication as well as PD-1 rs10204525 polymorphism, suggesting that increased PD-1 expression may affect the disease course of chronic HCV infection by facilitating HCV viral replication via suppressing host antiviral immune response, and this may at least partially relate to rs10204525 in PD-1 3’-UTR. These data may provide immunogenetic information for monitoring disease prognosis and designing immunotherapeutic approaches to HCV-associated diseases.
Acknowledgements
The authors would like to acknowledge all patients for their participation in this study.
Funding
This work was supported by funding from the Institut Pasteur du Maroc
Experimental Ethics
All individuals gave informed consent and the study was approved by the Ethics Committee of The Faculty of Medicine of Casablanca in accordance with the ethical guidelines of the Declaration of Helsinki
Disclosure of Conflicts of Interest The authors declare no conflicts of interest
References
- Shepard CW, Finelli L, Alter MJ (2005) Global epidemiology of hepatitis C virus infection. Lancet Infect Dis 5: 558–567. [crossref]
- Averhoff FM, Glass N, Holtzman D (2012) Global Burden of Hepatitis C: Considerations for Healthcare Providers in the United States. Clin Infect Dis 55 suppl_1: S10–5. [crossref]
- Grebely J, Dore G (2011) What Is Killing People with Hepatitis C Virus Infection. Semin Liver Dis 31: 331–339. [crossref]
- Kittlesen DJ, Chianese-Bullock KA, Yao ZQ, Braciale TJ, Hahn YS, et al. (2000) Interaction between complement receptor gC1qR and hepatitis C virus core protein inhibits T-lymphocyte proliferation. J Clin Invest 106: 1239–1249. [crossref]
- Yao ZQ, King E, Prayther D, Yin D, Moorman J, et al. (2007) T Cell Dysfunction by Hepatitis C Virus Core Protein Involves PD-1/PDL-1 Signaling. Viral Immunol 20: 276–87. [crossref]
- Sharpe AH, Wherry EJ, Ahmed R, Freeman GJ (2007) The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat Immunol 8: 239–245. [crossref]
- Ishida Y, Agata Y, Shibahara K, Honjo T (1992) Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J 11: 3887–3895. [crossref]
- Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, et al. (2009) PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med 206: 3015–3029. [crossref]
- Keir ME, Butte MJ, Freeman GJ, Sharpe AH (2008) PD-1 and Its Ligands in Tolerance and Immunity. Annu Rev Immunol 26: 677–704. [crossref]
- Dellgren C, Gad HH, Hamming OJ, Melchjorsen J, Hartmann R, et al. (2009) Human interferon-?3 is a potent member of the type III interferon family. Genes Immun 10: 125–131.
- Penna A, Pilli M, Zerbini A, Orlandini A, Mezzadri S, et al. (2007) Dysfunction and functional restoration of HCV-specific CD8 responses in chronic hepatitis C virus infection. Hepatology 45: 588–601. [crossref]
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, et al. (2006) Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439: 682–687. [crossref]
- Radziewicz H, Ibegbu CC, Fernandez ML, Workowski KA, Obideen K, et al. (2007) Liver-infiltrating lymphocytes in chronic human hepatitis C virus infection display an exhausted phenotype with high levels of PD-1 and low levels of CD127 expression. J Virol 81: 2545–2553. [crossref]
- Takayama T, Sekine T, Makuuchi M, Yamasaki S, Kosuge T, et al. (2000) Adoptive immunotherapy to lower postsurgical recurrence rates of hepatocellular carcinoma: a randomised trial. Lancet (London, England) 356: 802–807. [crossref]
- Yoong KF, McNab G, Hübscher SG, Adams DH (1998) Vascular adhesion protein-1 and ICAM-1 support the adhesion of tumor-infiltrating lymphocytes to tumor endothelium in human hepatocellular carcinoma. J Immunol 160: 3978–3988. [crossref]
- Shi F, Shi M, Zeng Z, Qi R-Z, Liu Z-W, et al. (2011) PD-1 and PD-L1 upregulation promotes CD8+ T-cell apoptosis and postoperative recurrence in hepatocellular carcinoma patients. Int J Cancer 128: 887–896. [crossref]
- Xiao W, Zhang Q, Deng XZ, Jiang LF, Zhu DY, et al. (2015) Genetic variations of IL-28B and PD-1 are in association with the susceptibility and outcomes of HCV infection in Southeast China. Infect Genet Evol 32: 89–96. [crossref]
- Li Z, Li N, Zhu Q, Zhang G, Han Q, et al. (2013) Genetic variations of PD1 and TIM3 are differentially and interactively associated with the development of cirrhosis and HCC in patients with chronic HBV infection. Infect Genet Evol 14: 240–246. [crossref]
- Hua Z, Li D, Xiang G, Xu F, Jie G, et al. (2011) PD-1 polymorphisms are associated with sporadic breast cancer in Chinese Han population of Northeast China. Breast Cancer Res Treat 129: 195–201. [crossref]
- Haghshenas MR, Naeimi S, Talei A, Ghaderi A, Erfani N, et al. (2011) Program death 1 (PD1) haplotyping in patients with breast carcinoma. Mol Biol Rep 38: 4205–4210. [crossref]
- Mojtahedi Z, Mohmedi M, Rahimifar S, Erfani N, Hosseini SV, et al. (2012) Programmed death-1 gene polymorphism (PD-1.5 C/T) is associated with colon cancer. Gene 508: 229–32. [crossref]
- Savabkar S, Azimzadeh P, Chaleshi V, Nazemalhosseini Mojarad E, Aghdaei HA, et al. (2013) Programmed death-1 gene polymorphism (PD-1.5 C/T) is associated with gastric cancer. Gastroenterol Hepatol from bed to bench 6: 178–182. [crossref]
- Tang W, Chen S, Chen Y, Lin J, Lin J, et al. (2017) Programmed death-1 polymorphisms is associated with risk of esophagogastric junction adenocarcinoma in the Chinese Han population: A case-control study involving 2,740 subjects. Oncotarget 8: 39198–3208. [crossref]
- Qiu H, Zheng L, Tang W, Yin P, Cheng F, et al. (2014) Programmed death-1 (PD-1) polymorphisms in Chinese patients with esophageal cancer. Clin Biochem 47: 612–617. [crossref]
- Zhang G, Li N, Zhang P, Li F, Yang C, et al. (2014) PD-1 mRNA expression is associated with clinical and viral profile and PD1 3'-untranslated region polymorphism in patients with chronic HBV infection. Immunol Lett 162: 212–216. [crossref]
- Wu S-C, Chang SC, Wu H-Y, Liao P-J, Chang M-F, et al. (2008) Hepatitis C virus NS5A protein down-regulates the expression of spindle gene Aspm through PKR-p38 signaling pathway. J Biol Chem 283: 29396–29404. [crossref]
- Axley P, Ahmed Z, Ravi S, Singal AK (2018) Hepatitis C Virus and Hepatocellular Carcinoma: A Narrative Review. J Clin Transl Hepatol 6: 1–6. [crossref]
- Ringelhan M, McKeating JA, Protzer U (2017) Viral hepatitis and liver cancer. Philos Trans R Soc Lond B Biol Sci 372. [crossref]
- Ezzikouri S, Benjelloun S, Pineau P (2013) Human genetic variation and the risk of hepatocellular carcinoma development. Hepatol Int 7: 820–831. [crossref]
- Park HJ, Park JS, Jeong YH, Son J, Ban YH, et al. (2015) PD-1 Upregulated on Regulatory T Cells during Chronic Virus Infection Enhances the Suppression of CD8+ T Cell immune response via the Interaction with PD-L1 Expressed on CD8+ T Cells. J Immunol 194: 5801–5811. [crossref]
- Rutebemberwa A, Ray SC, Astemborski J, Levine J, Liu L, et al. (2008) High-programmed death-1 levels on hepatitis C virus-specific T cells during acute infection are associated with viral persistence and require preservation of cognate antigen during chronic infection. J Immunol 181: 8215–8225. [crossref]
- Kasprowicz V, zur Wiesch JS, Kuntzen T, Nolan BE, Longworth S, et al. (2011) High Level of PD-1 Expression on Hepatitis C Virus (HCV)-Specific CD8+ and CD4+ T Cells during Acute HCV Infection, Irrespective of Clinical Outcome. J Virol 85: 4633–4633. [crossref]
- Golden-Mason L, Klarquist J, Wahed AS, Rosen HR (2008) Cutting edge: programmed death-1 expression is increased on immunocytes in chronic hepatitis C virus and predicts failure of response to antiviral therapy: race-dependent differences. J Immunol 180: 3637–3641. [crossref]
- Chihab H, Jadid F-Z, Foka P, Zaidane I, El Fihry R, et al. (2018) Programmed cell death-1 3'-untranslated region polymorphism is associated with spontaneous clearance of hepatitis B virus infection. J Med Virol 90: 1730-1738.[crossref]
- Baba H, Kettani A, Bouqdayr M, Ouladlahsen A, Bensghir R, et al. (2021) Programmed cell death-1 single-nucleotide polymorphism rs10204525 is associated with human immunodeficiency virus type 1 RNA viral load in HIV-1-infected Moroccan subjects. Med Microbiol Immunol 210: 187-196. [crossref]
