Resistiogram Pattern of Escherichia Coli Isolated from Various Clinical Samples In & Around Kanchipuram
S Sivasankari1*, S Senthamarai1, C Anitha1, K Muthulakshmi1, VM Somasunder1, K Akila1, J Subha1, Sijimol1
1 Meenakshi Medical College & Research Inistitute, Enathur, Kanchipuram, Tamil Nadu, India.
*Corresponding Author: S Sivasankari, Meenakshi Medical College & Research Inistitute, Enathur, Kanchipuram, Tamil Nadu, India, TEL: 044 2726 1096; FAX: 044 2726 1096; E-mail:Murugansivasankari1@gmail.com
Citation: S Sivasankari, S Senthamarai, C Anitha, K Muthulakshmi, VM Somasunder, et al. (2019) Resistiogram Pattern of Escherichia Coli Isolated from Various Clinical Samples In & Around Kanchipuram. Allergy drugs clin immunol 3:116.
Copyright:© 2019 S Sivasankari, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Received date: September 05, 2019; Accepted date: September 19, 2019; Published date: September 24, 2019.
Introduction
Escherichia coli is one of the important cause of nosocomial infections in humans [1]. E.coli is widely implicated in various clinical infections as hospital acquired and community acquired [2]. Pathogenic isolates of E.coli have relatively high potentials for developing resistance. Resistance to gram negative bacteria have great concern as these organisms are becoming resistant to Penicillin, cephalosporin & monobactums [3]. Increasing bacterial resistance is now highly prevalent in developing countries and is currently worrisome problem. Antimicrobial resistance is a growing threat worldwide with different resistant mechanisms [4]. The production of Extended spectrum of beta lactamase is an important mechanism which is responsible for the resistance to 3rd generation cephalosporin [5]. Among the wide array of antibiotics β lactams are widely used agents accounting for over 50% of all systemic antibiotics in use [6]. The most common cause of resistance to β lactam antibiotics is the production of β lactamases. ESBL are plasmid borne and confer multiple drug resistance making the infection severe and difficult to treat [7]. In India ESBL, E.coli ranges from 41 – 60% highest is 86% reported in 2017. Fluroquinolone antibiotics are now continued to increase, which is of great concern, which accounts 20% of HAI [8]. β lactam resistance, fluroquinolone resistance with bacterial biofilm is considered as a pathogenic treat for Nosocomial infections . E.coli is a common human pathogen being MDR, including resistance to quinolones [9]. There is always a gap in the knowledge based and this makes the patient care largely empherical. This makes physicians to prescribe multiple antibiotics which is both cost and morbidity wise increase. In order to curb the problem, some primary data on the resistance pattern is to be known for selecting appropriate drugs. Bacterial biofilm has been considered as a virulence factor contributing to infections associated with various medical devices, causing nosocomial infection [10]. The tendency of micro organism to develop biofilm has been well documented for number of medical devices. This process is particularly relevant because biofilm associated micro organisms are much more resistant to antimicrobial agents. E.coli being one of the commonest organism causing hospital acquired infections exhibits ESBL production which renders organism resistant to β lactam group of drugs leading to limited treatment options. In our hospital, E.coli is one of the common organism encountered everyday in routine culture reporting. Hence this study done to know the prevalence of ESBL E.coli, their resistance pattern and their virulence factors.
Aim & Objectives
To study the prevalence of bacterial resistance in relation to biofilm forming E.coli from various clinical samples. To do of follow of the study for 2 yrs.
Objectives: To isolate and identify E.coli from various clinical sample. To study the drug resistance among the E.coli isolates by Kirby bauer disc diffusion method. To study the biofilm formation in relation to drug resistance. To study the ESBL & MBL production among E.coli isolates.
Material & Methods
Source of data: Meenakshi Medical College hospital & Research Institute. Ethical committee approval was obtained. Study period: Sep 2017 to Dec 2017. Sample size: Sample included were Urine, Sputum, Pus & wound swab. Samples of urine, sputum, pus & swabs were all collected as per standard precautions & aseptically [11]. Identification done based on colony Morphology, staining series of biochemical tests as per standard protocol [12]. Antibiogram done by kirby bauer disc diffusion method as per CLSI guidelines for Gentamicin(10µg), Norfloxacin(10µg), amikacin(30µg), nitrofurantoin (300µg), nalidixic acid(30µg), cotrimaxazole(25µg), amoxyclave(30µg), ofloxacin(5µg), levofloxacin(5µg), ceftazidime(30µg), cephotaxime(30µg), ciprofloxacin(10µg), imipeneme10µg), ceftriaxone(30μg), Cefepime(30μg) [13]. If the isolate were resistant to 5 or more classes of drugs, it is taken as multidrug resistant. MIC done for the highest resistance of the drug. Detection of the biofilm was done by cong. Red agar method [14]. ESBL detection done by double disc diffusion method of ceftazidime and ceftazidime + clavulanic acid. Isolates resistance to carbapenems are screened for MBL. Imipenem – EDTA combined disc test done for MBL detection. MIC done for the Maximum resistant drug according to CLSI guidelines (ciprofloxcin≥4µg/ml).
Result
Total of 235 E.coli were isolated from 1522 samples, Among 235 E. coli, Urine isolation of E.coli is 129 (54.89%), Pus is 84 (35.74%) & Sputum is 22 (9.36%). Table 1.
Table 1: Table showing age & sex distribution of E.coli infection n=235.
| S.No | Age group | Male n=103 | Female n=132 | Total n=235 |
| 1. | 20 – 40 % | 34 (33.00%) | 86 (65.15%) | 120 (51.06%) |
| 2. | 41 – 60% | 23(22.33%) | 28 (21.21%) | 51 (21.70%) |
| 3 | > 60% | 46 (44.66%) | 18(13.63%) | 64(27.23%) |
E.coli isolated more in females of reproductive age group. Table 2.
Table 2: Table showing Antimicrobial resistance pattern of E.coli isolates.
| Antimicrobial agents | Resistance (n) | Resistance (%) |
| Nalidixic acid (30µg) | 119 | 50.6% |
| Cofrimoxazole (25µg) | 98 | 41.7% |
| Ciprofloxacin (10µg) | 135 | 57.6% |
| Amoxyclav(30µg) | 121 | 51.48% |
| Norfloxacin (10µg) | 83 | 35.31% |
| Ceftazidime (30µg) | 148 | 62.97% |
| Cephotaxime (30µg) | 148 | 62.97% |
| Cetriaxone (30µg) | 145 | 61.70% |
| Gentamicin (10µg) | 124 | 52.76% |
| Oflaxacin (5µg) | 130 | 55.31% |
| Cefapime (30µg) | 71 | 30.21% |
| Piperacillin(100μg ) | 32 | 13.6% |
| Imipenem (10µg) | 21 | 8.9% |
Maximum resistance seen in 3rd generation Cephalosporin followed by Ciprofloxacin. Table 3.
Table 3:
| MDR Strains | 169 | 46.38% |
169 strain were Multidrug resistant. Apart from ESBL screening drugs, Ciprofloxacin is next highest resistant drug.
The MIC of ciprofloxacin ranged from 8 – 64 µg /ml.
The breakpoint by MIC value ≥ 4 µg/ ml were defined as resistant isolates as per CLSI guidelines.
Table 4,5. 29 isolates were strong biofilm producers and were multidrug resistant.
Table 4: Resistance mechanisms (ESBL & MBL) of E.coli isolates.
| Resistance Mechanism | No.of positives | % |
| ESBL | 53 | 22.55% |
| MBL | 1 | 1.27% |
Table 5: Biofilm formation by congo Red agar method done for E.coli isolates.
| N | % |
| 29 | 12.34 |
Discussion
In our study total of 235 E.coli were isolated from 1522 samples. E.coli isolated from urine, 56% (13.2 )were predominant, followed by pus 34% (32) &9.2%(22) sputum. Many studies done in India, Aishwarya et al 2018 showed the E.coli predominant isolation from urine samples [20]. In our study, in the age distribution, more number of isolates seen in reproductive age group female preponderance is more. This could be due to the changes in vaginal pH during pregnancy. Similar studies done by, sanjo et al 2017 [21]. In our study E.coli showed maximum resistance to nalidixic acid(50.6%) followed by cotrimaxozole (41.7%) similar studies done by sanjo et al 2017 [21]. In our study Gentamicin resistance is 52.76% this is slightly lower with zohreh et al 2018 who also reported 62.25% resistance to Gentamicin. Fluroquinolone resistance varies from region to region [4]. The highest Fluroquinolone resistance were reported to be 75% in UTI infection [15]. MDR is pervasive and a growing clinical problem, which is a threat and increasing economic burden due to over usage of antibiotics [16]. Nosocomial E.coli are more common nowadays, because of MDR pattern is raising [17]. In our study 169 (46.38%) were found to be MDR. This is concordant with a study done by Aishwarya et al 2018 also reported 43.12% of MDR E.coli. In our study 53 (22.55%) isolates were ESBL positive and 1 isolate was MBL positive similar findings were also reported kaur J etal [18,19]. Overall prevalence of ESBL producer were found to vary greatly in different geographical areas and in different institutes. Several studies in India have reported ESBL production varying from 20% to 84% [21]. Among the 169 MDR strains 29 (12.34%) were found to be strong biofilm produces similar studies done by Jayanthi Ray et al 2015 showed 14.2% of E.coli were biofilm produces & all were MDR E.coli [22].
Summary
A total of 235 E.coli isolates were isolated from various clinical samples. (Urine, Pus, Sputum). Majority of E.coli isolates were from urine (169) followed by pus (84). High prevalence of E.coli infection was seen in females (132) than Males (n=103). Percentage of isolation were seen more in age group of 20 – 40 yrs in females & more than 60 yrs in males. Antibiotic susceptibility testing showed a high resistance of ciprofloxacin 57.61% and also for 3rd generation Cephalosporin 62.97%. Minimal resistance encountered for Imipenem (8.9%) and hence still the drug of choice. The 135 ciprofloxacin resistant strains were subjected to MIC by agar dilution method and based on the interpretation of CLSI breakpoints were in the range of 8 to > 32µg/ml. Among 235 isolates, 169 (46.38%) were multidrug resistant.53 (22.55%) were ESBL producers & 1 were MBL producer. In our study biofilm producers were found to be 29 (17.34%). Ability of biofilm formation of clinical strains is the major virulence determinal of E.coli and results demonstrate that the presence of E.coli in harsh conditions may develop more pathogenic potential due to genetic changes and gene transfer mechanisms [14].
Conclusion
The present study clearly highlights that ESBL producers were resistant to 3rd generation Cephalosporin but still sensitive to Imipenem. In view of MDR E.coli, certain precautions were followed and the infection rate were decreasing. Antibiotic policy created and recycling of drugs were done routinely. Infection control strategy like disinfection of wards, barrier Precautions, compulsory hand washing after nursing care. Health education done to all patients & at tenders for prevention of infection. Ciprofloxacin resistance can be used as a surrogate marker of multidrug resistance, thus limiting already restricted treatment options.
Table 6: Follow up of study
| Year | No.of E.coli | MDR | Cipro R | ESBL | |||
|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | ||
| Jan - April 2018 | 248 | 112 | 46 .16 | 118 | 47.5 | 54 | 21.78 |
| May – Aug 2018 | 215 | 132 | 45.58 | 106 | 49.30 | 41 | 19.06 |
| Sep – Dec 2018 | 254 | 102 | 40.15 | 112 | 44.09 | 49 | 19.19 |
| Jan – April 2019 | 231 | 91 | 35.41 | 98 | 42.42 | 42 | 18.01 |
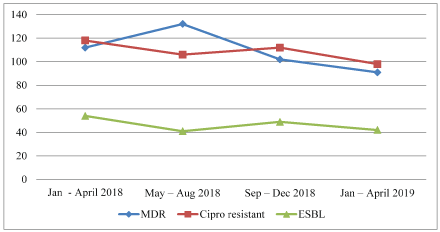
The MDR pattern, Ciprofloxacin resistance, ESBL E.coli were reducing with our strict infection control practices since Jan 2018 with our effective infection control practices like hand washing & wearing proper PPE & antibiotic policy , strict disinfection practices. So the infection rates have started to taper down since 1 year.
To Conclude
With the strict infection control strategies and by routine antibiotic surveillance & providing health education of standard precautions & infection control measures, the multidrug resistance controlled and prevented. The infection control team supervises the surveillance, Antimicrobial stewardship program, policy making & provided health education to all to minimize the infection.
References
- Mathavi Sureshkumar, Sasikala Gopinathan, Kondian Rangachari Rajesh, Indra Priyadharsini (2012) Prevalence of Ciprofloxacin Resistance Among Gram-Negative Bacilli in a Tertiary Care Hospital IDI: JCDR/2012/3490:1900
- Karlowsky JA, Jones ME, Draghi DC, Thornsbery C, Sahm DF, et al. (2004) Prevalence of antimicrobial susceptibilities of bacteria isolated from blood cultures of hospitalized patients in he United States in 2002. Ann.. Clin.Microbiol Antimicrob 3: 7.
- Curcio D (2013) Multidrug-resistant Gram-negative bacterial infections: are you ready for the challenge? Curr Clin Pharmacol. Mar 12. Cookson, A.L; Cooley W.A; Woodward, M.J (2002). The role of type 1and curly fimbriae of shiga toxin-producing Escherichia coli in adherence to abiotic surfaces. International Journal of Medical Microbiology 292: pp 195-120.
- Zohreh Aminzadeh, Davood Yadegarynia, Alireza Fatemi, Saieed Azad Armaki, Bahram Aslanbeygi, et al. (2013) Prevalence and Antimicrobial Susceptibility Pattern of Extended Spectrum Beta Lactamase (ESBL) and non-ESBL Producing Enteric Gram-Negative Bacteria and Activity of Nitrofurantoin in the era of ESBL. Jundishapur J Microbiol 6: e6699
- Umadevi S, Kandhakumari G, Joseph NM, Kumar S, Easow JM, et al. (2011) Prevalence and antimicrobial susceptibility pattern of ESBL producing Gram Negative Bacilli. Journal of Clinical and Diagnostic Research 5: 236-239.
- Shila Jalalpour (2012) Antibiogram pattern in extended spectrum beta lactamase nano enzyme producing gram negative bacilli in Iranian urinary tract infection African. journal of pharmacy and pharmacology 6: pp. 899-903.
- Sareaa MG Al-Mayahie (2013) Phenotypic and genotypic comparison of ESBL production by Vaginal Escherichia coli isolates from pregnant and non-pregnant women. Annals of Clinical Microbiology and Antimicrobials 12: 7.
- Ebbing Lautenbach, Joshua P Metlay, Xiangqun Mao, Xiaoyan Han, Neil O Fishman, et al. (2010) The Prevalence of Fluoroquinolone Resistance Mechanisms in Colonizing Escherichia coli Isolates from Hospitalized Patients. Clin Infect Dis 51: 280-285.
- Sam Bouchillon1, Daryl J Hoban, Robert Badal, Stephen Hawser (2012) Fluoroquinolone Resistance Among Gram -Negative urinary Tract Patho-gens: Global Smart Program Results, 2009-2010. The Open Microbiology Journal 6: 74-78.
- Lalit Meshram, Rakesh Kumar Patidar, Mayuri Khare, Swati Bagde, Keertisheel N Sahare, et al. (2012) Comparative analysis between biofilm formation of commensal and pathogenic Escherichia coli isolates Asiatic. Journal of Biotechnology Resources 03: 1401-1407.
- Bailey, Scott, Diagnostic Microbiology, 12th Edition, Lab cultivation and isolation of bacteria, 133 – 147
- Mackie, MacCartney. Practical Medical Microbiology. 14 Edition. Enterobacteriaceae, Echerichia, 363-364.
- CLSI (Clinical and Laboratory Standard Institutes 2017 Guidelines
- Sharma M Aparna, Yadav S, Chaudhary U (2009) Biofilm production in uropathogenic Escherichia coli. Indian J Pathol Microbiol 52: 294. [crossref]
- Niranjan V, Malini A (2014) Antimicrobial resistance pattern in Escherichia coli causing urinary tract infection among inpatients. Indian J Med Res. 139: 945-948.
- Neetu Sharma, Anita Gupta, Geeta Walia, Rupinder Bakhshi (2016) Pattern of Antimicrobial Resistance of Escherichia coli Isolates from Urinary Tract Infection Patients: A Three Year Retrospective Study. Journal of Applied Pharmaceutical Science 6: pp. 062-065.
- TekinA, AlD, Deveci O, Tekin R, Bozdag H, et al. (2012) In vitro efficacy of antibiotics in Escherichia coli strains isolated from Urine Cultures. The New Journal of Medicine 29: 88-91.
- Kaur J, Chopra S, Sheevani, Mahajan G (2013) Modified Double Disc Synergy Test to Detect ESBL Production in Urinary Isolates of Escherichia coli and Klebsiella pneumoniae. J Clin Diagn Res 7: 229-233.
- Mackie, Mccartney Practical Medical Microbiology 14th Edition Elsevier publication, Chapter 5 – Specimen collections, culture & Media p95 -111.
- Aishwarya Govindasamy, Vijeta Bajpai, Surabikurana Prevalence & charatisation of ß lactanen producing E.coli from teritary care hospital Journal of laboratory physicians 2019/vol 11/Issue 2/Pg123 – 127
- Sanjo Gupta, Veena Maheshwari, Rajesh (2017) Shahrevalence of ESBL Producing Escherichia coli and Klebsiella Species among Clinical Isolates and their in vitro Antimicrobial Susceptibility Pattern in a Tertiary Care Hospital. International Journal Current Microbiology Applied Science 6: 2295-2303.
- Jayanti Ray, Rudrajit Paul, Abhik Haldar, Sagnic Mondol (2015) A study on antibiotic resistance pattern of Escherichia coli isolated from urine specimens in Eastern India Open Access. International Journal Medical Science Public Health 4: 1670-1674.
